| |
Top 10 FishMicrosat Families |
|
 |
|
 |
 |
 |
 |
| | |
| |
Top 10 FishMicrosat Species |
|
 |
|
 |
 |
 |
 |
|
|
|
Repeats statistics The program finds the frequency of different type of repeats from the records available in FishMicrosat and displays in the tabular and pie chart form.
Maximum repeats types AC 19793 | TG 16047 | CA 15018
| | Repeats | Frequency | Repeats | Frequency | TGTCTG | 14 | GTGTGC | 10 | CCCTAA | 8 | CACACT | 8 | AGAAAG | 8 | GATGAA | 8 | AAAAAT | 6 | ATATAC | 6 | TTTTAT | 6 | AAAATA | 6 | CACACG | 6 | ACACAT | 6 | TTTATT | 4 | TAAATA | 4 | GAGGAA | 4 | CATCTT | 4 | AATCAG | 4 | CTTATT | 4 | GTGTCT | 4 | TACATA | 4 | AAGGAC | 4 | ACATAT | 4 | ATAATG | 4 | TATATG | 4 | GTGCGT | 4 | TAAAAA | 4 | AGAGAA | 4 | CCTGGT | 4 | CTAATG | 2 | TATAGT | 2 | TCACCA | 2 | AATATG | 2 | AGTTTA | 2 | GTGAAA | 2 | TTCTCT | 2 | GTACTT | 2 | AATCAT | 2 | GTGTGG | 2 | GAACTT | 2 | GAGTGC | 2 | ATAAAC | 2 | AATGTC | 2 | TTAGGG | 2 | CACGCA | 2 | GAAAGA | 2 | GAACCT | 2 | AGAACC | 2 | ACAAGA | 2 | TATATT | 2 | CTTCAT | 2 | GAGTAG | 2 | AATGGG | 2 | CATATA | 2 | ATACAT | 2 | TCTTTC | 2 | ACAGAG | 2 | CATACA | 2 | AGACAG | 2 | CTCGCA | 2 | AAAGAA | 2 | ATATTT | 2 | GAGATA | 2 | CCTAAC | 2 | TCTTAG | 2 | GGGGCT | 2 | TAATTT | 2 | GAATCT | 2 | CTTTGT | 2 | AGATAG | 2 | AAAGTG | 2 | GTCTGC | 2 | GTGGAA | 2 | GAAAGT | 2 | GCGTGT | 2 | GGAGAG | 2 | CCCCGT | 2 | GGATAG | 2 | CGATGG | 2 | ACCCTA | 2 | TCATTA | 2 | TTCAGA | 2 | GTCTCT | 2 | ACATAC | 2 | ATTTAC | 2 | GTAAAT | 2 | GCGGTG | 2 | TCTATC | 2 | CAGAAC | 2 | AATTAA | 2 | CTCTGT | 2 | GGAGGA | 2 | TGACTT | 2 | TGAATC | 2 | CAGACA | 2 | ATATAG | 2 | ATTCCC | 2 | GCTCCG | 2 | CATCAA | 2 | TATTTG | 2 | AGTTCA | 2 | ATAAAA | 2 | AGGAGA | 2 | TTTTTA | 2 | ATTTTT | 2 | CTCTCG | 2 | CAAGTA | 2 | CTAGTC | 2 | CATCGT | 2 | GAGAGT | 2 | CATTTA | 2 | CATCAC | 2 | TATTAA | 2 | TATACA | 2 | TCATCT | 2 | GTGTCC | 2 | TGTGCT | 2 | AGGGTT | 2 | TATTAG | 2 | TCCCTC | 2 | ATCAGG | 2 | TTAGCT | 2 | TCTGGT | 2 | AGGCAG | 2 | ATTTAT | 2 | TCTCTT | 2 | GTGAGT | 2 | CTCCTT | 2 | AAGCGT | 2 | CCAGCA | 2 | ATATAA | 2 | AGTCAG | 2 | CTCACT | 2 | AATAAC | 2 | ATACTT | 2 | AATTGT | 2 | AGCCGG | 2 | TAAATG | 2 | CGCACA | 2 | TCTCTG | 2 | TTTTCG | 2 | TAGGTT | 2 | GTGAGA | 2 | AGGACC | 2 | CAGCTG | 2 | CATTAT | 2 | ATAACT | 2 | ACACAG | 2 | AGGAAG | 2 | AAAACA | 2 | TATAGA | 2 | TTTGAA | 2 | TATGTG | 2 | TGAGGA | 2 | TCTATA | 2 | AGTGTA | 2 | GTCAAG | 2 | TGCCTC | 2 | TCAGAA | 2 | AGAAGG | 2 | GGGAGA | 2 | TGCGGT | 2 | AAACTT | 2 | TTTATA | 2 | CAGGAC | 2 |
| |
| This pie chart repersents maximum frequencies of all types (mono, di, tri, tetera, penta and hexa) nucleotide repeats. 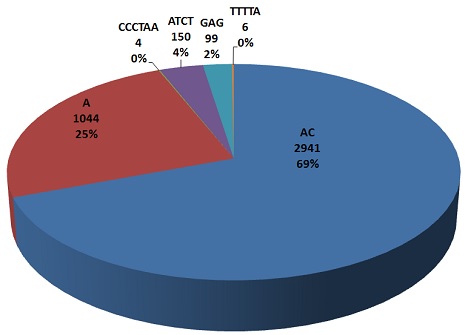
|
|
|