| |
Top 10 FishMicrosat Families |
|
 |
|
 |
 |
 |
 |
| | |
| |
Top 10 FishMicrosat Species |
|
 |
|
 |
 |
 |
 |
|
|
|
Repeats statistics The program finds the frequency of different type of repeats from the records available in FishMicrosat and displays in the tabular and pie chart form.
Maximum repeats types AC 9896 | TG 8022 | CA 7508
| | Repeats | Frequency | Repeats | Frequency | TTTTA | 70 | TATTA | 64 | AAAAT | 63 | ATAAT | 58 | AAATA | 57 | ATTAT | 54 | TAAAA | 54 | ATATA | 49 | TAATA | 45 | TTTAT | 44 | AATAA | 41 | TATAT | 40 | ATTTT | 38 | TATTT | 38 | AAATT | 37 | TTATT | 37 | ATATT | 35 | TTATA | 35 | TATAA | 34 | AATAT | 29 | ATAAA | 29 | TATTC | 24 | TTCTA | 23 | AGAAT | 22 | TAGAA | 21 | TTTTG | 21 | ATTCT | 20 | AAAAC | 20 | AATAG | 18 | GAATA | 18 | TTATG | 16 | TCTAT | 16 | TTTAA | 15 | ATTAA | 15 | AATTA | 14 | TTTCT | 13 | ATTAG | 12 | CTATT | 12 | TTAAA | 11 | ATAGA | 11 | AAGAA | 10 | ATTTA | 10 | CAAAA | 10 | CTTTT | 10 | GAAGA | 9 | TTTAG | 9 | TTTGT | 9 | TTAGA | 9 | GTTTT | 9 | AGAGA | 8 | TTGAA | 8 | AATCT | 8 | TTTTC | 8 | ACAAA | 8 | AGAGG | 8 | TCTCT | 8 | AAAAG | 8 | AATTT | 8 | AAAGA | 8 | AAACA | 7 | GAGAA | 7 | TAGTA | 7 | TAAAT | 7 | ATAAC | 7 | TGTTA | 7 | GAAAA | 7 | AAGTC | 7 | TTGTT | 7 | TCTTC | 6 | AACAT | 6 | TACTA | 6 | CTCTT | 6 | ATTCA | 6 | TATGT | 6 | CTCTC | 6 | AGAAA | 6 | TTAAT | 6 | ATATG | 6 | TTCTT | 5 | AACAA | 5 | AGTAT | 5 | TTGTA | 5 | ATGTT | 5 | GAGGA | 5 | TTCTC | 5 | TCAAG | 5 | AACTG | 5 | ATATC | 5 | TCTTT | 5 | TTTCA | 5 | ATTTC | 4 | TCAAT | 4 | ATCTA | 4 | TATAC | 4 | GTTAT | 4 | TAATT | 4 | AGATT | 4 | AAAGG | 4 | AGGTT | 4 | TAGTT | 4 | AACTA | 4 | CATAT | 4 | CCTTT | 4 | TGACT | 4 | GACTT | 4 | GATAT | 4 | ATAGT | 4 | AGAAG | 4 | TCTCC | 4 | ACGAC | 4 | TTTGC | 3 | TTATC | 3 | GTTCA | 3 | TGAAT | 3 | TCCTC | 3 | CAAAT | 3 | AACAC | 3 | GATGA | 3 | TGTTT | 3 | TTCTG | 3 | GTGGT | 3 | AGTCA | 3 | AATTG | 3 | GAGAG | 3 | AGGAA | 3 | CTTCT | 3 | AAATG | 3 | ACATA | 3 | AAGAG | 3 | CAAGT | 3 | CAAAG | 3 | TGTTC | 3 | AAACT | 3 | ACAAC | 3 | ACTAA | 3 | ATGTA | 3 | TTACA | 2 | TGTGT | 2 | TAAAC | 2 | CTTGC | 2 | TATGA | 2 | TCTCA | 2 | TGATA | 2 | GTTAC | 2 | CAGAC | 2 | CTAAA | 2 | AGGAG | 2 | TATTG | 2 | TTAGT | 2 | TGCTT | 2 | AGGCA | 2 | GGGAA | 2 | TCAAA | 2 | TAATC | 2 | TGAAG | 2 | TCAAC | 2 | AAGTG | 2 | TCCTT | 2 | ACAGG | 2 | GATGT | 2 | GTTTA | 2 | AATGT | 2 | ATACT | 2 | AATGA | 2 | ATGAA | 2 | AGATG | 2 | TCATG | 2 | CTTTA | 2 | TTGAC | 2 | ATCAC | 2 | GGAGA | 2 | ATTTG | 2 | TATAG | 2 | GATTT | 2 | TAAGA | 2 | CTATA | 2 | GTGAA | 2 | GAACA | 2 | CTCCT | 2 | AAATC | 2 | GAATG | 2 | CACTT | 2 | AATCA | 2 | CTGTT | 2 | AGACA | 2 | AGACG | 2 | TAATG | 2 | CTTTC | 2 | ACGTG | 2 | AGTTT | 2 | TATGG | 2 | CCTCT | 2 | CATTG | 2 | TATCG | 1 | ACGGC | 1 | AGATC | 1 | TTCCT | 1 | AGCCG | 1 | ATGGA | 1 | CCTAT | 1 | TAGGA | 1 | TGGTG | 1 | CTATC | 1 | ACGTA | 1 | AGTGT | 1 | CTAAC | 1 | TCGTG | 1 | GGAGG | 1 | GAACC | 1 | AGGAC | 1 | GAGGC | 1 | TATCA | 1 | GTGAC | 1 | CAGGA | 1 | ATAGG | 1 | GAATT | 1 | ACACA | 1 | AGCAG | 1 | AAGCT | 1 | AAAGC | 1 | GCTCG | 1 | TTACC | 1 | GTGTT | 1 | CAGAA | 1 | TTTGA | 1 | CTCAA | 1 | AACCT | 1 | TTGAT | 1 | AACCA | 1 | CCTCA | 1 | AATTC | 1 | ATTGT | 1 | GTTCT | 1 | ATTGA | 1 | TCTGG | 1 | TAGAT | 1 | CACAA | 1 | CACAT | 1 | GACAT | 1 | TGTTG | 1 | ATGAC | 1 | CTCCA | 1 | TCTTG | 1 | GAAGG | 1 | GCGGG | 1 | ATCAT | 1 | AGACT | 1 | GGACT | 1 | CTGTC | 1 | TAACG | 1 | CTCAC | 1 | CTGAA | 1 | GTATC | 1 | TGTCC | 1 | ACACT | 1 | TCGTA | 1 | ACACG | 1 | AGTTC | 1 | AAGAT | 1 | ACCAG | 1 | TTGCC | 1 | ATGAT | 1 | GGCAA | 1 | TCTAA | 1 | GCCTT | 1 | TGATT | 1 | AGTTG | 1 | TGTGC | 1 | CAATT | 1 | CTCTA | 1 | CCCCG | 1 | CCCCT | 1 | AAGGC | 1 | TACGT | 1 | ATGCA | 1 | CAAGG | 1 | TTAGC | 1 | TAGAG | 1 | GATCT | 1 | CAAAC | 1 | ACCGA | 1 | GATAA | 1 | GCCGG | 1 | GAGAC | 1 | CGTCG | 1 | ATCCA | 1 | GGTTC | 1 | TATGC | 1 | TCTGT | 1 | TGAGT | 1 | TACTG | 1 | ACAGA | 1 | GATTA | 1 | GGGAC | 1 | TTGCT | 1 | GTAAA | 1 | AACAG | 1 | AAGGA | 1 | TGGAG | 1 | AGGGA | 1 | TGCTG | 1 | TACTC | 1 | GGGAG | 1 | CATTA | 1 | TAACA | 1 | TCCAT | 1 | GATAC | 1 | GAGAT | 1 | GGCAG | 1 | TACCT | 1 | ATGGC | 1 | ATTAC | 1 | AAAGT | 1 | TACCC | 1 | ATGCT | 1 | GTATT | 1 | AATAC | 1 | TGCAA | 1 | TAAGT | 1 | CGTCT | 1 | AGAAC | 1 | TGTAT | 1 | TAGGC | 1 | GACGT | 1 | TTCAA | 1 | TCATT | 1 | ATGTC | 1 | GATGC | 1 | GCCTG | 1 | GATCA | 1 | AAGCA | 1 | GTGTA | 1 | TTTCC | 1 | GTCAA | 1 | TTCAT | 1 | TTACT | 1 | CCAGT | 1 |
| |
| This pie chart repersents maximum frequencies of all types (mono, di, tri, tetera, penta and hexa) nucleotide repeats. 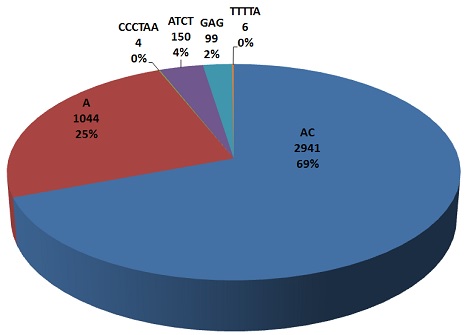
|
|
|